The definitive map of cancer resistance.
Cancer care is still too often a high-stakes guess. Enso is building a causal AI platform that learns from histopathology, molecular profiles, and clinical context — and then tests the biology to turn insight into action.
“The secret of the care of the patient is in caring for the patient.”
Francis W. Peabody — a reminder that technology must serve humanity.
A platform built for actionability
Precision oncology needs more than prediction. It needs robust generalization, mechanistic grounding, and a workflow that respects both doctor and patient.
Multimodal foundation
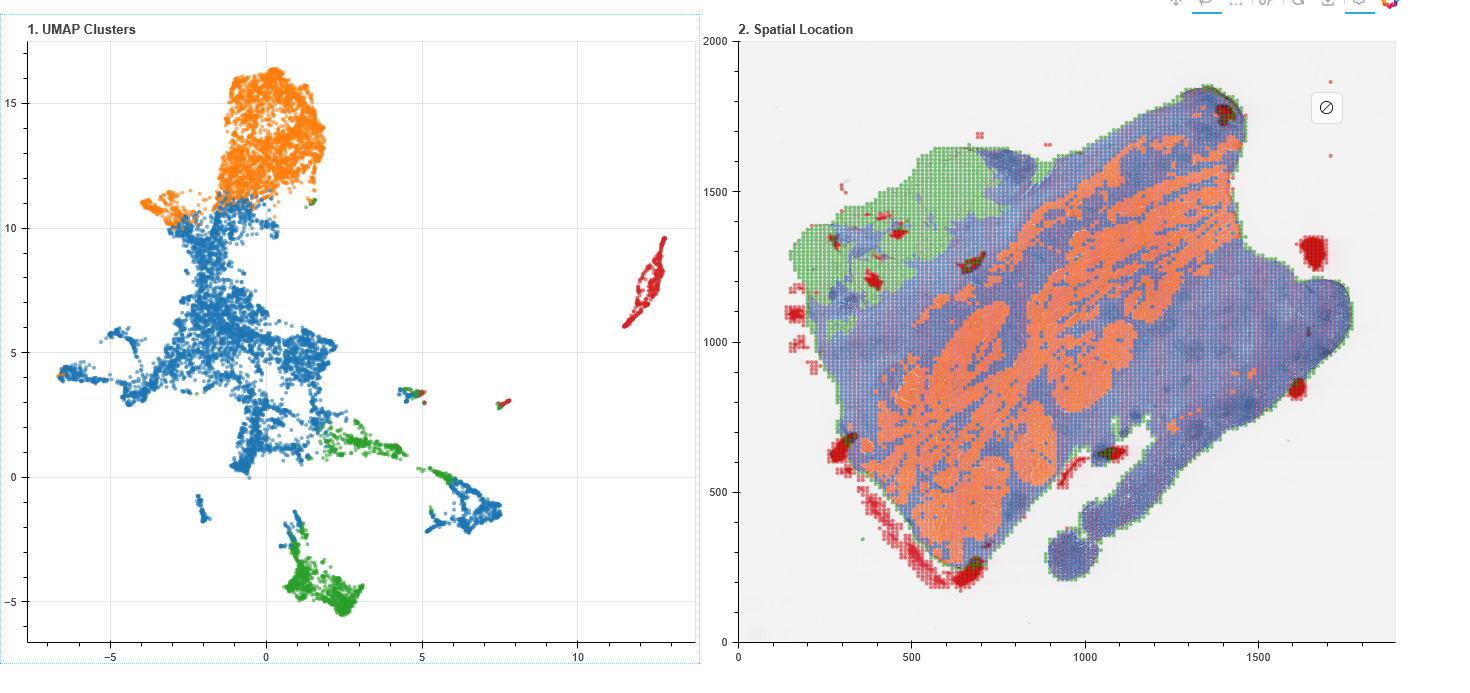
We start with the richest signal: tissue. We then align histology with transcriptomics and proteomics, and later with clinical narrative and exposure history — so the representation learns what matters.
Causal validation loop
Our models propose resistance mechanisms and biomarkers; we validate key hypotheses in the wet lab (e.g., functional genomics screens), closing the loop from correlation to cause.
Clinician-centered
The goal is not to replace clinicians — it’s to give them time back, reduce uncertainty, and offer transparent evidence in a domain where decisions are rarely deterministic.
Roadmap
We build from robust foundations toward prospective validation. Each step is designed to produce measurable learning — and better care.
Histology foundation model
Train a long-context slide model on public cohorts (TCGA, GTEx, CPTAC), evaluate on external benchmarks, and build the infrastructure that makes scaling inevitable.
Proteomics-aware learning
Add a proteomic regression objective on CPTAC to enrich the latent space with molecular signal. This is the first step from pattern recognition to actionable biology.
Validation and pilots
Partner with leading cancer centers for pilot studies and prospective evaluation; validate key mechanisms with functional assays; translate insights into clinical and trial decision support.
A global map — for everyone
Resistance mechanisms vary across populations. We want to build with the world, not just for it — increasing representation, reducing bias, and returning value to partner institutions globally.
Partnerships
We work with hospitals, cancer centers, researchers, and biotech to build models that are robust, validated, and useful.
Hospitals & cancer centers
Collaborate on data and validation. Partners can be recognized in publications, receive early access, and help shape a system designed for real workflows.
- De-identified slide + outcome cohorts, with clear governance.
- External validation and site-specific robustness analysis.
- Priority access / partner pricing for pilots.
Pharma & biotech
Use the causal engine to de-risk trials, identify biomarkers, and uncover resistance mechanisms worth validating.
- Patient stratification and biomarker discovery.
- Mechanism hypotheses grounded in multimodal evidence.
- Validation strategy aligned with wet-lab experimentation.
Team
We are an interdisciplinary founding group spanning medicine, machine learning, and experimental biology.
Luca Maria Menga
Founder · MD/BSc (Humanitas University & Politecnico di Milano, MEDTEC). Research experience across digital pathology, structural biology, and high-throughput wet lab (Harvard, Stanford/SLAC).
Matteo
Cofounder · Building scalable systems and product foundations for clinical-grade deployment.
Mattia
Cofounder · Physics & statistics background. Focused on rigor, evaluation, and modeling strategy.
Contact
If you are a cancer center, hospital, research group, or biotech team and want to collaborate, we would love to talk.