From tissue to causality
Our platform is designed around a simple principle: models should not just predict — they should earn trust. That requires robustness, interpretability, and experimental validation.
The core object: a patient embedding
We learn a low-dimensional representation that captures the essential biology of a patient’s tumor — a space where similar mechanisms cluster together, and where differences are measurable rather than anecdotal.
- Histology for architecture, microenvironment, and morphology.
- Transcriptomics / proteomics to ground representations in molecular function.
- Clinical context to connect biology to real outcomes and treatment choices.
Why “low-dimensional” matters
Medicine is overwhelmed by complexity, but complexity is not the same as randomness. If we can uncover the underlying structure, we can act with more clarity — and more humility.
How it works
A concrete, modular pipeline — built so each part can be evaluated and improved independently.
1) Ingest
Whole-slide images, molecular profiles, and structured clinical context are harmonized into a single training substrate. We start with public cohorts (TCGA, GTEx, CPTAC) and progressively add proprietary partner datasets.
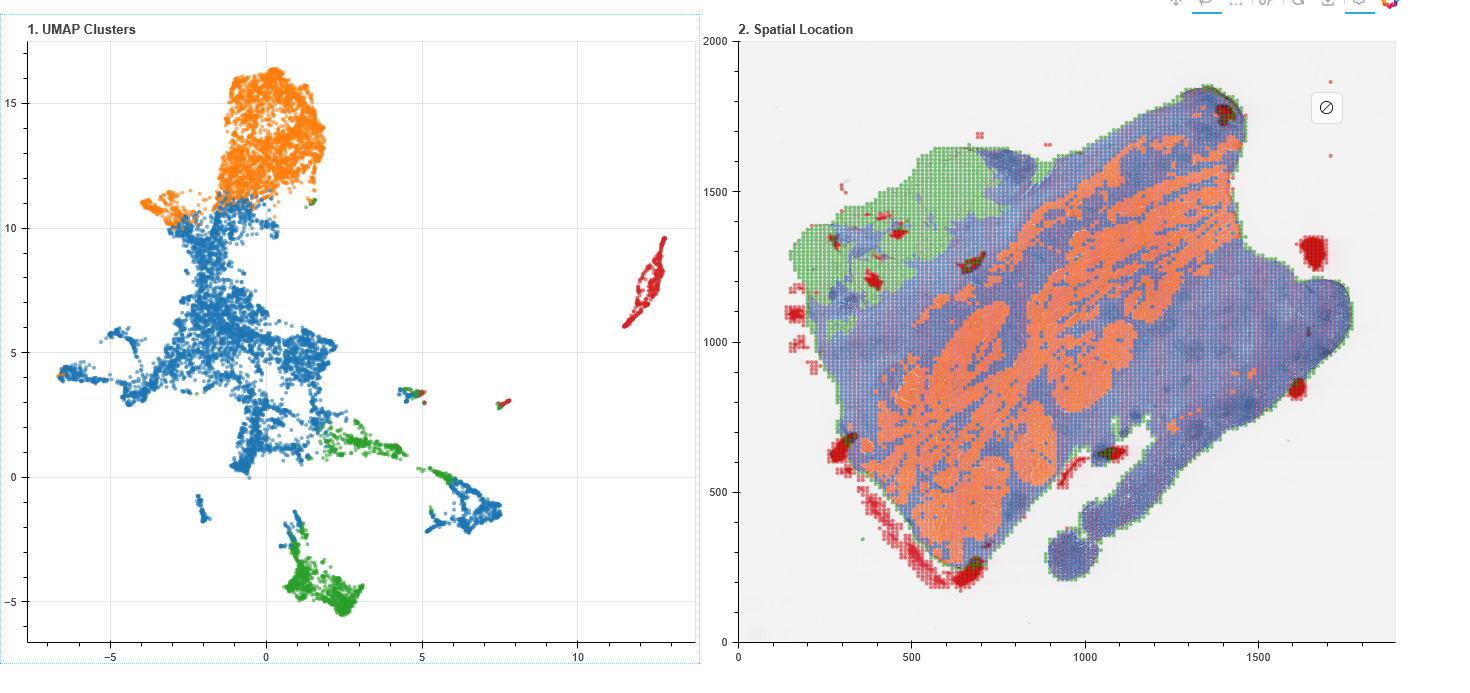
2) Represent
We encode tissue from local cellular texture to long-range spatial structure. Long-context modeling is essential: many clinically meaningful patterns emerge at scale.
3) Align
We align histology with transcriptomic and proteomic signals so the representation becomes “molecularly aware” — not just visually descriptive.
4) Predict
Downstream heads translate the embedding into clinically relevant tasks: response prediction, stratification, biomarker hypotheses, and uncertainty estimates.
5) Validate
Top hypotheses can be tested with functional genomics and wet-lab experiments. Validation converts correlations into mechanisms.
6) Deploy
The user interface must respect clinical reality: auditability, interpretability, and workflow integration — designed to give clinicians time back.
What we will not do
We’re building with ambition, but also with discipline.
- No clinical claims without validation. We do not present research outputs as medical advice.
- No black boxes. Every model must be stress-tested across hospitals and cohorts.
- No “AI as a slogan.” We treat modeling, datasets, and lab work as a single scientific system.